Pfam and peptide coordinates
In the table of peptide binding Pfams, select a Pfam, ProtCid returns a table listing all structures containing the Pfam and peptides.
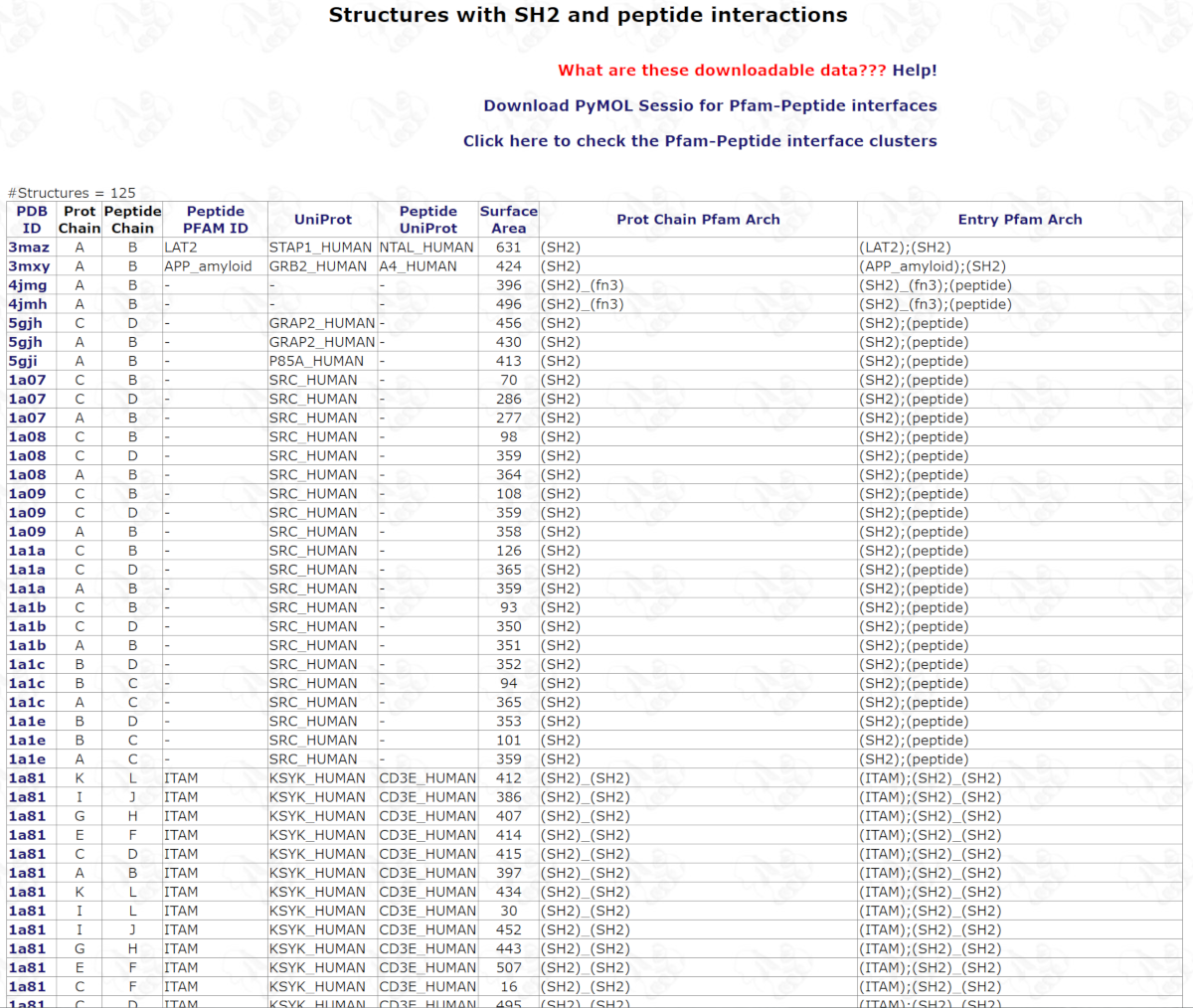
In the structure web page for the Pfam like Pfam SH2 ,
click "Download coordinates of domains, peptides and PyMOL scripts" to download the coordinates of all domains, peptides and four PyMOL script files, named by Pfam ID_pep.tar.gz (SH2_pep.tar.gz).
The Pfam domains are aligned either based on default model of PyMOL (_byChain.pml and _byDomain.pml) or based on Pfam HMM positions (_pairFit.pml and pairFitDomain.pml).
_byDomain.pml and _pairFitDomain.pml display just the Pfam domains, while _byChain.pml and _pairFit.pml display whole chains including Pfam domains and other segments if there are.
Pfam domains are colored in rainbow, from red to blue, from N to C terminus. The segments other than domains are colored in light grey.
Some domains may not be aligned well first. Manually aligning them is necessary by PyMOL command like "align 1aaa, 1bbb".
1bbb is the first domain file in a PyMOL script file, it is the center domain. All other domains are aligned to it.
The following picture shows the Pymol figure of SH2 domains and peptides which are aligned by SH2 domains.
The interactions of Pfam domains and peptides are not clustered.
The color of SH2 domains is changed to green (PyMol command: "color green, chain A" ),
and the color of peptides is changed to magenta (PyMol command: "color magenta, chain B").
Pfam-peptide interface clusters
Click the "Click here to check the Pfam-Peptide interface clusters" link to retrieve the Pfam-peptide interface clusters
and download coordinates and sequences for each cluster.
The first cluster of SH2 and peptide interfaces
contains 123 structures, 34 different proteins (UniProt codes).
The minimum sequence identity of protein sequences is 9%. Buried surface area is 442.
Like chain and domain interface cluster page, the coordinates and sequences of clusters can be downloaded.
| Column Name |
Annotation |
| Cluster ID |
An integer ID (1:N) |
| #Entries |
the number of PDB entries |
| #UniProt |
the number of Uniprots of Pfam domains |
| #UniProt(Peptide) |
the number of Uniprots of peptides |
| #Pfams(Peptide) |
the number of Pfams of peptides |
| MinSeqID |
the minimum sequence identity of Pfam domains |
| SurfaceArea |
the accessible surface area |
Structures of each cluster can be viewed by click the "+" sign. The following picture shows the structures of the first cluster of SH2-peptide interfaces.